- Received On: 2022-07-24|
- Accepted On: 2022-11-17|
- Published On: 2022-12-03
| Article Metadata | |||
|---|---|---|---|
| 1 | Submitted Manuscript | PPD/MIN/22213/1 | |
| 2 | Cover Letter to Editor | PPD/CLE/22213/1 | |
| 3 | Copyright Transfer Letter | PPD/CTL/22213/1 | |
| 4 | Authors’ Consent Letter | PPD/ACL/22213/1 | |
| 5 | Initial Editorial Screening Report | PPD/IESR/22213/1 | |
| 6 | Review Agreement Letter (Reviewer 1) | PPD/RAL/22213/R1 | |
| 7 | Review Agreement Letter (Reviewer 2) | PPD/RAL/22213/R2 | |
| 8 | Manuscript Review Report (Round 1, Reviewer 1) | PPD/MRR/22213/R1.1 | |
| 9 | Manuscript Review Report (Round 1, Reviewer 2) | PPD/MRR/22213/R1.2 | |
| 10 | Revised Manuscript | PPD/MIN/22213R | |
| 11 | Review Response Letter (Round 1) | PPD/RRL/22213/R1 | |
| 12 | Manuscript Review Report (Round 2, Reviewer 1) | PPD/MRR/22213/R2.1 | |
| 13 | Manuscript Review Report (Round 2, Reviewer 2) | PPD/MRR/22213/R2.2 | |
| 14 | Final Editorial Screening Report | PPD/FESR/22213 | |
| 15 | Letter of Acceptance and Acknowledgement | PPD/LAA/22213 | |
| 16 | Accepted Manuscript | PPD/MIN/22213A | |
| Request Access | |||
| Supplementary Data | ||
|---|---|---|
| The datasets used and/or analyzed during the study are available from the corresponding author on reasonable request. | ||
| Datasets | Not Provided | Request Availability |
Abstract
Background: Polymicrobial wound infection has been a matter of concern because of the rapid spread of antibiotic-resistant strains. An attempt was made to evaluate the prevalence and antibiotic susceptibility pattern of pathogens isolated from clinically suspected wound-infected patients.
Methods: Pathogens found in swabs and pus was identified by their cultural, morphological, and microscopic characteristics, while antibiotic susceptibility patterns were determined using the modified Kirby-Bauer disc diffusion method.
Results: A total of 374 swab samples were taken, with 330 (88.23%) showing bacterial growth, among which 221 (67%) were found to be gram negative. Pathogenic bacteria found in this study included Pseudomonas spp. (43%), Staphylococcus aureus (23.30%), Escherichia coli (10.30%), Enterococcus faecalis (9.70%), Klebsiella pneumoniae (7.90%), and Proteus mirabilis (5.80%). Meropenem was found to be the most sensitive antibiotic, while cefixime and clindamycin had the worst effectiveness against most pathogens (exceptionally, Proteus mirabilis showed 100% resistance to Cephradine).
Conclusion: This study implicated 16 antibiotics on 330 isolates, and on average, 12 drugs showed moderate to high resistance patterns. This could be attributed to indiscriminate antibiotic usage, poor handling of diagnostic laboratory instruments and services, and a lack of appropriate drug selection guidelines. The data generated by the study illuminates the importance of responsible use of medications.
Significance Statement
The study was not only of scientific interest but also of social value. It revealed that people belong to age group of 21-40 years and male were the most vulnerable affected population in this study. Among the identified pathogen Pseudomonas spp. was most prevalent. Meropenem was the most efficient whereas cephalosporin antibiotics were least efficient antibiotics found in the study. As a cross-sectional and localized study, it may play a role for comparative analysis which can add new information in medical field.
Introduction
Following a loss of skin integrity (i.e., a wound), subcutaneous tissue is exposed, providing a moist, warm, and nutritive environment for bacteria colonization and proliferation [1]. A variety of microbial and host variables like bacterial inoculums, virulence factors, conditions of the microenvironment of the wound, etc. are thought to have a role in the progression of a wound from uninfected to infected [2]. A wide range of microorganisms, including bacteria, fungi, parasites, and viruses, can infect a wound [3]. For S. aureus, P. aeruginosa, E. coli, Klebsiella spp., and Acinetobacter spp. are the most prevalent organisms reported previously [4-6].
Antibiotics, on the other hand, have been proven to be quite useful in the treatment and prevention of infections. The value of antibiotics in minimizing wound infections has been clearly defined by the timing of administration, type of antimicrobial agent, and duration of administration. However, antibiotic resistance development has emerged as a remarkable threat to the elimination of wound infection [7].
Wound infection causes sepsis, limb loss, extended hospital stays, and increased costs, and is responsible for major human mortality and morbidity around the world [8]. Previous investigations by Soltani et al., 1998 indicated that Pseudomonas aeruginosa-originated burn infection mortality was raised by up to 40–50% [9]. However, it's one of the most prevalent infections picked up in hospitals [10]. In hospitals located in the rural areas of Bangladesh, the scenario of personal hygiene of hospital staff is not satisfactory. Moreover, pathological medical waste materials originating from various sources such as diagnostic equipment (syringes, broken bottles, blood and urine bags, gloves) are poorly handled and not properly managed [11]. This poor handling and mismanagement may lead to the dispersal of wound-infecting pathogens in hospitals. Although it is a recurring problem and can’t be eradicated completely, it can be reduced to a minimum by taking immediate control measures against the most commonly isolated organism and using good wound care [12]. Hence, the current study was conducted to track the prevalence of the causative agents of wound infection as well as the antibiotic susceptibility pattern of the isolated pathogens.
Methods
Obtaining Samples
A total of 374 wound swabs were collected from clinically suspected wound-infected patients in different hospitals in Tangail district, Bangladesh from June to December, 2020. During sample collection, the Levine technique, i.e., wiping the infected area with 0.9% sodium chloride followed by swabbing of clean tissue [13]. A total of 330 wound swab culture positive cases were selected from 374 swab samples for further microbiological investigation.
Identification of positive swab culture isolates
Each sample was inoculated on MacConkey agar, Hi-Chrome agar, Blood agar, Chocolate agar, Eosin Methylene Blue agar, and Mannitol Salt Agar in the laboratory and incubated for 24-48 hours at 37°C to ensure the growth of specific pathogenic bacterial flora. However, the morphological, microscopic, and biochemical properties of these bacteria were determined to identify them with the help of Bergey’s Manual of Determinative Bacteriology [14,15].
Antibiotic susceptibility testing
The Kirby-Bauer disc diffusion method was performed for this test (antibiotic susceptibility testing). For the sensitivity test of the test strains, fourteen commercial antibiotics (Oxoid, UK) were used. The commercial antibiotic discs used for the study were Amoxicillin (10µg), Azithromycin (15µg), Cefixime (05µg), Cephradine (30µg), Ceftriaxone (30µg), Cefuroxime (30µg), Ciprofloxacin (05µg), Clindamycin (2µg), Cefepime (30µg), Gentamycin (10µg), Meropenem (10µg), Nitrofurantoin (300µg), Levofloxacin (05µg), Trimethoprim (5µg). Amikacin (12µg) and Flucloxacillin (18µg). These antibiotics were selected due to their wide range of usage in our subject area which will be helpful to reflect the usual antibiotic sensitivity and resistance pattern of the pathogen infecting local area. This test was carried out by streaking isolated pathogens on Muller Hinton Agar plate and incubating them at 37°C for 24 hours. The "zone of inhibition" was assessed after incubation and classified as sensitive, moderate, or resistant according to CLSI standards [16].
Data Analysis
Prevalence of bacteriological isolates involved in wound infection were analyzed by using Microsoft excel, 2019. Moreover, by using these tools, antibiotic susceptibility and resistance pattern was determined. Since, antimicrobial resistance was done once, so implementation of statistical analysis such as: standard error and deviation was excluded.
Results
Demographical study of wound infected patient
Age-based stratification of patients with wound infections
A total of 330 patients were selected for an infectious disease study and categorized into different age-groups of people as follows (Figure-1). Among them, the 21–40 age group of people was the highest in number (138 patients, 41.8%), followed by <20 years (83 patients, 25.1%), 41–60 years (78 patients, 23.7%) and > 61 years (31 patients, 9.4%).
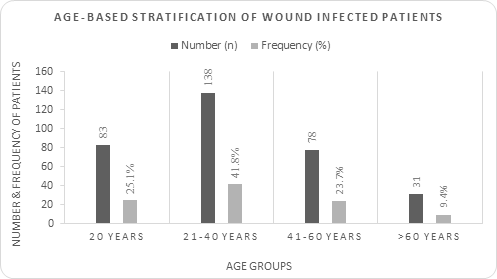
Figure 1: Age-based distribution of wound infected patients
Age groups are placed in X-axis and, respective frequency and number is placed in Y-axis. Blue bars indicating number whereas red bars indicating frequency. These frequencies were determined by dividing number of patients of specific age groups by total number of patients. Therefore, the value indicated in red bar can be used as relative abundance and compared to each other.
Gender-based distribution of wound-infected patients
Wound-infected people of different classes can again be distinguished by gender-based parameters. Figure 2 shows that the number of male patients (173 patients, 52.4%) was greater than the number of female patients (157 patients, 47.6%) due to random sampling.
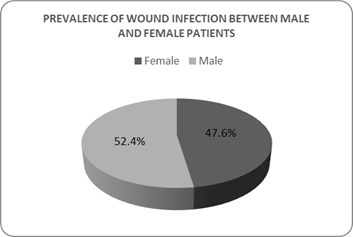
Figure 2: Gender-based distribution of wound infected patients
Total patients (n)=330, Male patient= 173 (52.4%, indicated in red colour), Female patient= 157 (47.6%, indicated in blue colour). These frequencies were determined by dividing number of patients of specific gender groups by total number of patients. Therefore, the value indicated in red and blue portion of pie chart can be used as relative abundance and compared to each other.
Prevalence of bacterial pathogens isolated from patients with wound infections
From 374 clinically suspected wound-infected patients, 330 wound swab culture positive cases (88.23%) were isolated from wound swab and pus. After identification, E. coli, Enterococcus faecalis, Klebsiella pneumoniae, and Proteus mirabilis were isolated. Pseudomonas aeruginosa and Staphylococcus aureus were confirmed. Among them, Pseudomonas aeruginosa (142 isolates, 43%) was the most frequently found virulent pathogen, followed by Staphylococcus aureus (77 isolates, 23.30%), E. coli (34 isolates, 10.30%), Enterococcus faecalis (32 isolates, 9.70%), Klebsiella pneumoniae (26 isolates, 7.90%), and Proteus mirabilis (19 isolates, 5.80%) Figure 3. In total, Gram negative bacteria accounted for 221 (67%) of the 330 bacterial isolates, whereas Gram positive bacteria accounted for 109 (33%) of the total in this study.
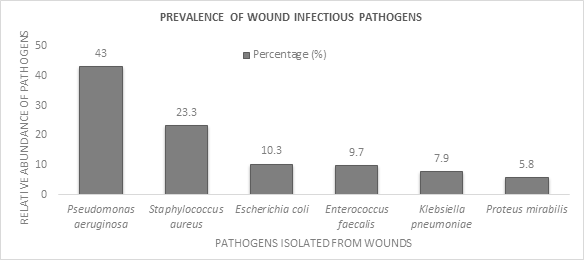
Figure 3: Prevalence of pathogen isolated from wound swab and pus sample of diseased patients
All of the isolates’ frequency represented here by their respective abundance in percentage (blue bar). The value was determined by dividing the number of specific pathogens by total number of pathogens and used to compare with the abundance of other pathogens.
Biochemical parameters for bacterial identification
The identification of the pure bacterial isolates was performed by biochemical parameters as presented in Table 1. In case of P. aeruginosa provided positive results in motility test, oxidase and catalase test but the urease test result was somewhat variable. In case of E. coli exhibiting both positive and negative test results producing strains were found in gas production, motility and indole test medium. All E. coli isolates showed positive test results in catalase, citrate and MR test and negative results in VP. These isolates could not metabolize Urea and failed to produce hydrogen sulfide. K. pneumoniae exerted similar mechanisms in KIA but differed in the result of MIU medium in view of the utilization of urea. P. mirabilis, S. aureus, E. faecalis all of three isolates oxidase negative and catalase positive. However, E. faecalis showed negative results in all cases except citrate and vp test.
Table 1: Results of biochemical parameters of the isolates
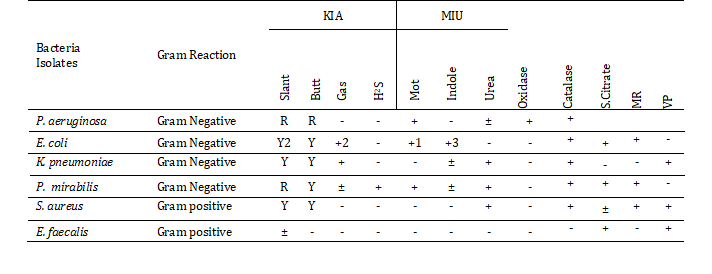
Note: (+) =Positive; (-) =Negative reaction; (±) =Variable; R=Red (Alkaline reaction); Y=Yellow (Acid reaction); KIA=Kliger's Iron Agar; MIU= Motility Indole Urea Agar; H2S (+) =Hydrogen sulfide (Blackening); 1=A few strains are non-motile; 2= A few strains produce red-pink slant; 3= A minority strains give a negative result; Mot=Motility test, catalase =Catalase test, S.Citrate=Simmons citrate test, MR=Methyl red test, VP=Voges -Proskauer test.
The biochemical test results of these isolated pathogens were compared with the standard description of Bergey’s manual of Determinative Bacteriology 8th edition [9].
Antibiotic susceptibility profiling of isolated pathogens
Pseudomonas aeruginosa was found to be the most prevalent pathogen in our study. The pathogen showed maximum sensitivity against Meropenem (97.2%) and no sensitivity against Cefixime. Like Pseudomonas aeruginosa, all isolates showed maximum sensitivity against Meropenem, but the resistance pattern was different. For example, 98.7% of sensitive cases were reported against Meropenem by Staphylococcus aureus and 94.7% of sensitive cases were found by Proteus mirabilis. The remaining isolates were completely sensitive to Meropenem. Pseudomonas aeruginosa and E. coli showed 99.3% and 100% resistance against Cefixime, respectively, which was the maximum for corresponding bacterial pathogens. In contrast, other isolates like Enterococcus faecalis (96.9%), Staphylococcus aureus (100%), Klebsiella pneumoniae (100%) and Proteus mirabilis (100%) exerted the highest frequency rate of resistant cases against Clindamycin. In addition, Proteus mirabilis and E. coli showed 100% resistance against cephradine and Clindamycin. Similarly, all isolates showed the highest intermediate responses to ciprofloxacin (Table 2).
Table 2: Antimicrobial sensitivity pattern of wound infectious pathogens. Frequency of sensitivity, intermediate and resistance was showed by percentage.
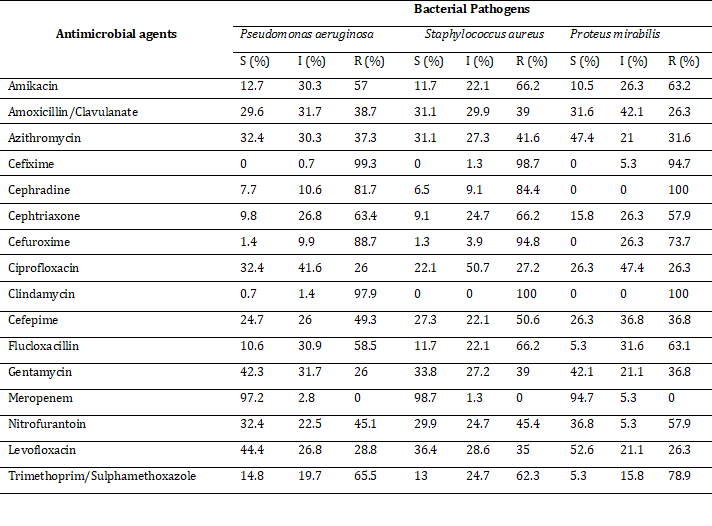
Note: S=Sensitive, I=Intermediate, R=Resistance, data presented in percentage.
Discussion
Wound colonization involves a variety of bacteria, e.g., synergistic aerobic, facultative, and anaerobic microbes, that can cause wound infections [17,18] and are resistant to multiple antibiotics. This draws attention to the need for scientists and practitioners to focus on polymicrobial wounds (often through combination interactions) and their treatment strategies. From this perspective, 374 wound-infected patients were selected for an epidemiological study, and 330 culture-positive samples were found from them. The remaining 44 samples were found to be negative for several reasons, such as sampling from the infected person practicing antibiotic therapy [19], absence of bacterial infection [20] and the failure of the recovery of the pathogen [20]. Gram-negative bacteria were found to be the most common (67%) in our investigation, which agrees with Yakha et al. ,2014; Abdullah et al., 2020; Abedin et al., 2020; Abedin et al., 2022 [16,21-23]. According to the authors, Gram-negative bacteria were isolated more frequently in this study because they are more prevalent among aerobes and facultative anaerobes in abscesses and skin wounds. Rai et al., 2017 opined that the presence of S. aureus on non-living objects, health professionals, and diseased people is the most potential source of its higher frequency in hospital-borne wound infected patients [24]. That is why other Gram-positive bacteria are not routinely found in wound-infected patients.
For swab sample collection, there were more male than female patients, which could be attributable to males' higher involvement in physical work for a living as opposed to females, as well as the increased risk of injuries during the activities. This study found that male patients (52.4%) had more growth positive cases than female patients (47.6%), which were corroborated by comparable studies conducted by KC et al., 2013 [8].
According to Mahat et al., 2017 the age range of 21–40 years accounted for 41.8 percent (138/330) [19]. In this study, we found that a high percentage of this age group is involved because there might be active participation in various physical and mechanical activities. Moreover, in rural areas, these age groups are more vulnerable due to their lifestyle, type of work and so are admitted to hospital in more numbers than the older people. As sample collection was performed from hospitals randomly, seriously injured and infected patients were mostly young. In rural areas, elderly people are rarely seriously injured or killed as a result of physical or mechanical work. As a result, in our study, older people were found to be less prone to wound infection than younger people.
Pseudomonas spp. were found to be the most common (43%), followed by S. aureus (23.4%), E. coli (10.3%), Enterococcus faecalis (9.7%), Klebsiella pneumoniae (8.0%), and Proteus mirabilis (5.6%). Similar findings were observed by Masaadeh and Jaran, 2009 [25] and Ranjan et al., 2010 [26]. They found Pseudomonas spp. were the most common bacteria among the total cases, accounting for 27.8% and 29.6% of the total cases, respectively. According to Zafar et al., 2007 [27] Pseudomonas spp. were the second most prevalent bacteria with 18.35 %, while S. aureus was the most common isolate with 41.28% of the total cases. Positive cultures found from infected sites do not nullify the probability of other contaminant bacteria such as Pseudomonas spp. [28]. Levine’s technique provided the best solution regarding sample collection from wounds where the word can be excluded without contamination [28]. For identification purposes, clinical features of wound infections and cultural characteristics of Pseudomonas spp. were taken into consideration. Therefore, there is a minimum possibility that isolated growths are not contaminants and absolutely pure growth.
In this study, Meropenem was found to be the most effective antibiotic against both Gram-negative and Gram-positive isolates. However, in a study by Yakha et al., 2014 [23], it was found that Amikacin was the most effective medicine for treating both Gram-negative and Gram-positive bacteria, which happened to be a considerably less sensitive antibiotic in our investigation. All pathogens were found to be most resistant to Cefixime and Clindamycin. Other antibiotics showed significant cases of intermediate and resistance responses towards the wound infection isolates. Staphylococcus aureus showed intermediate sensitivity towards Ciprofloxacin (50.7%) and the highest resistance towards Clindamycin (100%), which is supported by Ahmadishooli et al., 2020 [29]. They also reported that Methicillin-resistant S. aureus strains are resistant to Clindamycin, whereas Erythromycin-resistant germs exhibit a proclivity for Clindamycin resistance after therapy. People in rural areas use antibiotics without a doctor’s prescription and do not complete the full dose of antibiotic. Moreover, antibiotics such as Cefixime are taken by people without having knowledge regarding the nature and sensitivity pattern of the pathogen [30]. Thus, indiscriminate uptake of antibiotics may lead to resistance to Clindamycin and Cefixime. The development of penicillinase enzymes that hydrolyze the β–lactam ring in S. aureus is linked to β–lactam antibiotic resistance. The manufacture of PBP2a, a penicillin-binding protein located on the bacterial cell wall, is primarily responsible for MRSA resistance to β -lactam antibiotics. The bacterium can survive and multiply because it has a low affinity for β-lactams. 12 of the 16 antibiotics employed in this investigation demonstrated ineffectiveness, indicating that S. aureus isolates were from the MRSA and MDR families. However, it exhibited the maximum sensitivity to Meropenem (100%) and intermediate responsiveness to Ciprofloxacin (50.7%), which is comparable to Al-Mugdadi's findings [31], but there is a little discrepancy about the use of Amikacin as an effective antibiotic. Vancomycin and Teicoplanin, glycopeptide antibiotics, are viable alternatives. This study also revealed that E. coli is the third most common bacteria, accounting for 10.30% of all isolates and exerting significant cases of resistance towards β–lactam antibiotics (especially cephalosporin antibiotics), with the exception of Meropenem. So, it can be predicted that those were ESBL strains. They did, however, show 100% sensitivity to Meropenem, followed by Ciprofloxacin (50%), and Levofloxacin (47%), which could be a drug of choice for synergistic applications. The effectiveness of Meropenem against E. coli infection is also indicated in the study of Shaikh et al., 2015 [32]. Similar results were observed in Klebsiella pneumoniae where Meropenem, and Levofloxacin were the most effective and intermediate antibiotics, respectively. In addition, resistance to β–lactam drugs by multiple bacteria was also reported in the study of Ahmed, 2020 [21]. In our research work, Enterococcus faecalis and Proteus mirabilis showed similar types of sensitivity and resistant patterns as other isolated pathogens found in our research work, which is supported by other studies [33].
A cumulative antimicrobial strategy has already been observed in a variety of cases. Clindamycin and Metronidazole success rates against anaerobic organisms are relatively low, but the pattern dramatically reverses when they are used in combination with an Aminoglycoside e.g., Gentamicin or a Cephalosporin e.g., Cefuroxime or Cefotaxime. Cephamycin or Cefoxitin has been routinely used as a single antibiotic for the treatment of established infections in the United States, but the development of novel antibiotic families such as Ureidopenicillins, Carbapenems, and β-lactam combos has opened up a new arena for effective therapeutic approach [21]. In this investigation, only one beta-lactam inhibitor, i.e., Clavulanate was attempted to explore the best antimicrobial therapeutic option. Other beta-lactam inhibitor drugs such as Tazobactam, Sulbactam are not available in our studied area and usually not prescribed by the doctors. Therefore, these beta-lactam inhibitory drugs, except for Clavulanate, do not reflect the usual antibiotic sensitive or resistant pattern of the pathogens infecting local people and therefore cannot be recommended as a therapeutic combination from a local perspective. Conventionally prescribed and mostly used antibiotics in this local area were considered in our study. Because most of our cultures revealed polymicrobial and multidrug resistant organisms, the overall antibiotic susceptibility was extremely variable.
Conclusion
Patients of different ages, both male and female, are exposed to communicable diseases throughout the world every year, but not all are at the same risk. In this research work, male patients and people in the age group of 21–40 was found to be the most vulnerable groups affected by wound infection. Significant numbers of bacterial pathogens, solely or combined, are involved in wound infection development. In our study, Gram-negative bacteria occupied the major portion, and Pseudomonas aeruginosa was mostly involved in it. However, the resistance of these bacterial pathogens against multiple drugs has now been a matter of concern throughout the world. We observed multiple drug-resistant bacteria against a group of antibiotics, including β-lactam antibiotics except Meropenem. Meropenem has been found as the most efficacious antibiotic, while Cefixime (a cephalosporin antibiotic) and Clindamycin were the most ineffective. We recommend that, antibiotic susceptibility profile of wound infectious pathogens of every patient should be determined prior to prescribing any antibiotic as it is not usually done by the people of rural areas. From our investigation, it is evident that antibiotics such as Amikacin (highest sensitivity 21.9%) and Amoxicillin/Clavulanate (highest sensitivity 34.6%) are working in a lower frequency of cases. Some antibiotics such as Azithromycin (highest sensitivity 47.4%), Ciprofloxacin (highest sensitivity 32.4%), Gentamycin (highest sensitivity 44.1%) and Levofloxacin (highest sensitivity 52.6%) still have some potential and are better to be used. Other antibiotics such as Flucoxacillin, Nitrofurantoin, Trimethoprim/Sulfamethoxazole, Cephra-dine, Ceftriaxone, and Cefuroxime have already been resistant significantly and may not be fruitful in the treatment of wound infections in the near future. Intermediate sensitivity against Meropenem has been observed at a lower frequency, which is alarming for the future. We suggest that patients suffering from multidrug resistance can be treated with Meropenem with non-Cephalosporin non β-Lactam antibiotic combinations after evaluating their susceptibility pattern.
Abbreviations
CLSI: Clinical and Laboratory Standards Institute; MRSA: Methicillin-resistant Staphylococcus aureus; MDR: Multidrug-resistant; PBP2a: Penicillin‐binding protein 2a.
Authors’ Contributions
This work was a collaborative effort between all authors. The study was conceived, coordinated, and supervised by MZA and AAA. MASM and NEKF conducted the experiments and created graphical displays. MASM, NEKF and MIS took part in the writing of the manuscript. FTZ, FA, SMM, and MEA contributed to the critical review and article editing. MNA and MBA contributed to the interpretation of data in order to obtain a scientific conclusion. The final manuscript was read and approved by the authors.
Acknowledgments
We would like to offer our deepest appreciation to Md. Sobur Mia, of the Lab Zone and Hormone Center, without whose permission and cooperation it would not be possible to collect samples. We are also thankful to all the laboratory personnel for their kind cooperation. This work was collaboration among all the authors in proper assistance and writing to conduct successful research.
Funding
This Study did not receive any specific grants from public, commercial, or non-profit funding bodies.
Availability of Data and Materials
The datasets used and/or analysed during this research are available upon reasonable request from the corresponding authors.
Ethics Approval and Consent to Participate
This study was approved by the Human Ethics Committee (HEC) from Khwaja Yunus Ali University (KYAU/HEC#2020/07). Authorization for access to participants who fit the inclusion criteria was also granted by the HEC. Participation in the study was on a completely voluntary basis, and all participants signed informed consent (KYAU/IC/20-07/01). All demographic information and blood samples were de-identified from the study team.
Consent for Publication
Not applicable.
Competing Interests
Before submitting the article, all authors were in accord and there were no conflicts of interest.
References
- Felgueiras HP, Amorim MT. Functionalization of electrospun polymeric wound dressings with antimicrobial peptides. Colloids and Surfaces B: Biointerfaces. 2017 Aug 1;156:133-48. DOI PubMed Google Scholar Link
- Fry DE, Fry RV. Surgical site infection: the host factor. AORN journal. 2007 Nov 1;86(5):801-14. DOI PubMed Google Scholar Link
- Xu Z, Hsia HC. The impact of microbial communities on wound healing: a review. Annals of plastic surgery. 2018 Jul 1;81(1):113-23. DOI PubMed Google Scholar Link
- Thanganadar Appapalam S, Muniyan A, Vasanthi Mohan K, Panchamoorthy R. A study on isolation, characterization, and exploration of multiantibiotic-resistant bacteria in the wound site of diabetic foot ulcer patients. The Inter J of Lower Extremity Wounds. 2021 Mar;20(1):6-14. DOI PubMed Google Scholar Link
- Premanath R, Suresh S, Alva PP, Akash SK. Biofilm forming abilities of microorganisms associated with diabetic wound infection: a study from a tertiary care hospital. Biomed. and Pharmacol. J. 2019 Jun 25;12(2):669-76. DOI Google Scholar Link
- Rebic V, Masic N, Teskeredzic S, Aljicevic M, Abduzaimovic A, Rebic D. The importance of Acinetobacter species in the hospital environment. Medical arc. 2018 Nov; 72(5):325. DOI PubMed PMC Google Scholar Link
- Walsh TL, Chan L, Konopka CI, Burkitt MJ, Moffa MA, Bremmer DN, Murillo MA, Watson C, Chan-Tompkins NH. Appropriateness of antibiotic management of uncomplicated skin and soft tissue infections in hospitalized adult patients. BMC Infectious Diseases. 2016 Dec;16(1):1-8. DOI PMC Google Scholar Link
- Kc R, Shrestha A, Sharma VK. Bacteriological study of wound infection and antibiotic susceptibility pattern of the isolates. Nepal j of sci and technol. 2013;14(2):143-50. DOI Google Scholar Link
- Soltani K, Zand R, Mirghasemi A. Epidemiology and mortality of burns in Tehran, Iran. Burns. 1998 Jun 1;24(4):325-8. DOI PubMed Google Scholar Link
- Gottrup F, Melling A, Hollander DA. An overview of surgical site infections: aetiology, incidence and risk factors. EWMA journal. 2005 Sep;5(2):11-5. Google Scholar Link
- Hassan MM, Ahmed SA, Rahman KA, Biswas TK. Pattern of medical waste management: existing scenario in Dhaka City, Bangladesh. BMC public health. 2008 Dec;8(1):1-0. Doi: 10.1186/1471-2458-8-36 DOI PubMed PMC Google Scholar Link
- Sivaram P, Sreekumar A. Preoperative factors influencing mortality and morbidity in peptic ulcer perforation. European J of Trauma and Emergency Surg. 2018 Apr; 44(2):251-7. DOI PubMed Google Scholar Link
- Stotts NA. Wound infection: diagnosis and management. In: Bryant R, Nix D, eds Acute and Chronic Wounds-E-Book. 2015 Dec 7:283. Link
- Buchanan RE, Gibbons NE. Bergey’s Manual of determinative Bacteriolog. 1974; 8th edn. Williams and Wilkins Co, Baltimore. Link
- Sari GL, Trihadiningrum Y. Isolation and identification of native bacteria from total petroleum hydrocarbon polluted soil in Wonocolo public oilfields, Indonesia. J of Ecolog. Eng. 2019;20(8). DOI Google Scholar Link
- Abedin MZ, Mia S, Helal HB, Sultana S, Barman SD, Yeasmin F, Arfat MdE4, Farnaz NEK, Koly FA, Khairujjaman Md, Sultana MA, Mukul MEH and Ahmed AA. Multi-Drug Resistance Patterns of Wound Causing Bacterial Infections in a Rural Hospital, Sirajganj, Bangladesh, Public H Open Acc. 2022; 6(1): 000196. DOI Link
- Choi Y, Banerjee A, McNish S, Couch KS, Torralba MG, Lucas S, Tovchigrechko A, Madupu R, Yooseph S, Nelson KE, Shanmugam VK. Co-occurrence of anaerobes in human chronic wounds. Microbial ecol. 2019 Apr;77(3):808-820. DOI PubMed Google Scholar Link
- Krüger W, Vielreicher S, Kapitan M, Jacobsen ID, Niemiec MJ. Fungal-bacterial interactions in health and disease. Pathogens. 2019 May 21;8(2):70. DOI PubMed Google Scholar Link
- Mahat P, Manandhar S, Baidya R. Bacteriological profile of wound infection and antibiotic susceptibility pattern of the isolates. J Microbiol Exp. 2017 Apr 17;4(5):126-33. DOI Google Scholar Link
- Bacterial wound culture. Testing.com. https://www.testing.com/tests/bacterial-wound-culture/. Published February 19, 2020. Accessed December 2, 2022. Link
- Ahmed AA, Juyee NA, Hasan SA and Abedin MZ. Microbiological Screening and Antimicrobial Sensitivity Profiling of Wound Infections in a Tertiary Care Hospital of Bangladesh, Euro. J. of Med. and Health Sci. 2020; 2(5):101-106. DOI Google Scholar Link
- Abedin MZ, Aktar MB, Zaman MSU, Jarin L, Mia AS, Abdullah AA, Hasan R, Khairujjaman M, Islam R, Uddin ME and Shilpi RY . The Predominance of nosocomial pathogens among patients with post-operative wound infections and evaluation of the antibiotic susceptibility patterns in rural hospitals in Bangladesh. Recent Adv Biol Med. 2020(a); 6(4): 9800005. Google Scholar
- Yakha JK, Sharma AR, Dahal N, Lekhak B, Banjara MR. Antibiotic susceptibility pattern of bacterial isolates causing wound infection among the patients visiting B & B hospital. Nepal j of sci and technol. 2014;15(2):91-6. DOI Google Scholar Link
- Rai S, Yadav UN, Pant ND, Yakha JK, Tripathi PP, Poudel A, Lekhak B. Bacteriological profile and antimicrobial susceptibility patterns of bacteria isolated from pus/wound swab samples from children attending a tertiary care hospital in Kathmandu, Nepal. Inter. j of microbiolog. 2017 Mar 6;2017. DOI Google Scholar Link
- Masaadeh HA, Jaran AS. Incident of Pseudomonas aeruginosa in post-operative wound infection. Am J Infect Dis. 2009;5(1):1-6. Link
- Ranjan KP, Ranjan N, Bansal SK, Arora DR. Prevalence of Pseudomonas aeruginosa in post-operative wound infection in a referral hospital in Haryana, India. J of lab physicians. 2010 Jul; 2(02):074-7. DOI PubMed PMC Google Scholar Link
- Zafar AI, Anwar NA, Ejaz HA. Bacteriology of infected wounds-A study conducted at Children Hospital Lahore. Biomedica. 2007 Jul;23(8):1-4. Google Scholar Link
- Cross HH. Obtaining a wound swab culture specimen. Nursing2020. 2014 Jul 1;44(7):68-9. DOI Google Scholar Link
- Ahmadishooli A, Davoodian P, Shoja S, Ahmadishooli B, Dadvand H, Hamadiyan H, Shahriarirad R. Frequency and antimicrobial susceptibility patterns of diabetic foot infection of patients from Bandar Abbas District, Southern Iran. J of patho. 2020 Jun 9;2020. DOI PubMed PMC Google Scholar Link
- Hamilton-Miller JM. Overview of cefixime use in community-acquired infections. Cli. Microbiol. and infect. 2000 Jan 1; 6:79-81. DOI PubMed Google Scholar Link
- Al-Mugdadi SF. Activity of meropenem and ciprofloxacin in vitro against oxacillin resistant Staphylococcus aureus. Al Mustansiriyah J of Pharmaceutical Sci. 2010 Dec 1;8(2):64-71. DOI Google Scholar Link
- Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Risk factors for acquisition of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae in North-Indian hospitals. Saudi J of Biol. Sci. 2015 Jan 1; 22(1):37-41. DOI PubMed PMC Google Scholar Link
- Baguma A, Musinguzi B, Kagirita AA, Bazira J. Antimicrobial resistance profile among bacteria isolated from patients presenting with wounds at Kabale Regional Referral Hospital, South western Uganda. 2020. Preprint. DOI Google Scholar Link
Update History
| Revision Number | Date | Details Of Changes |
|---|---|---|
| 1 | 2022-12-01 | Original Article; published at its accepted version (Reference Number: PPD/MIN/22213A). |
 Pharmacotherapy & Pharmascience Discovery
Pharmacotherapy & Pharmascience Discovery
